| The overall processes of ExCNVSS |
|
|
Our method can be summarized as follows: (1) It extracts base-level read coverage depth within each targeted exonic region from the read alignment data and merges them to generate concatenated base-level read coverage data. (2) To reduce the coverage bias effect, base-level read coverage data are normalized by our four-step normalization protocol. In each step, target exon read coverage data are considered to be evaluated from test data only or from the ratio of test and control data, according to the contents of the input, test data only, or both test and control data.
(3) The scale-space filtering is then applied to normalized base-level read coverage data using a Gaussian convolution for various scales according to a given scaling parameter.
By differentiating the scale-space filtered data twice and then finding zero-crossing points of the second derivatives, inflection points of the scale-space filtered data are calculated per scale. (4) Finally,the types and exact locations of CNVs of test data are obtained by using parametric baselines, which are evaluated from the normalized base-level coverage data, and by analyzing the finger print map, which is the collection of contours of the zero-crossing points for various scales.
|
|
|
| |
|
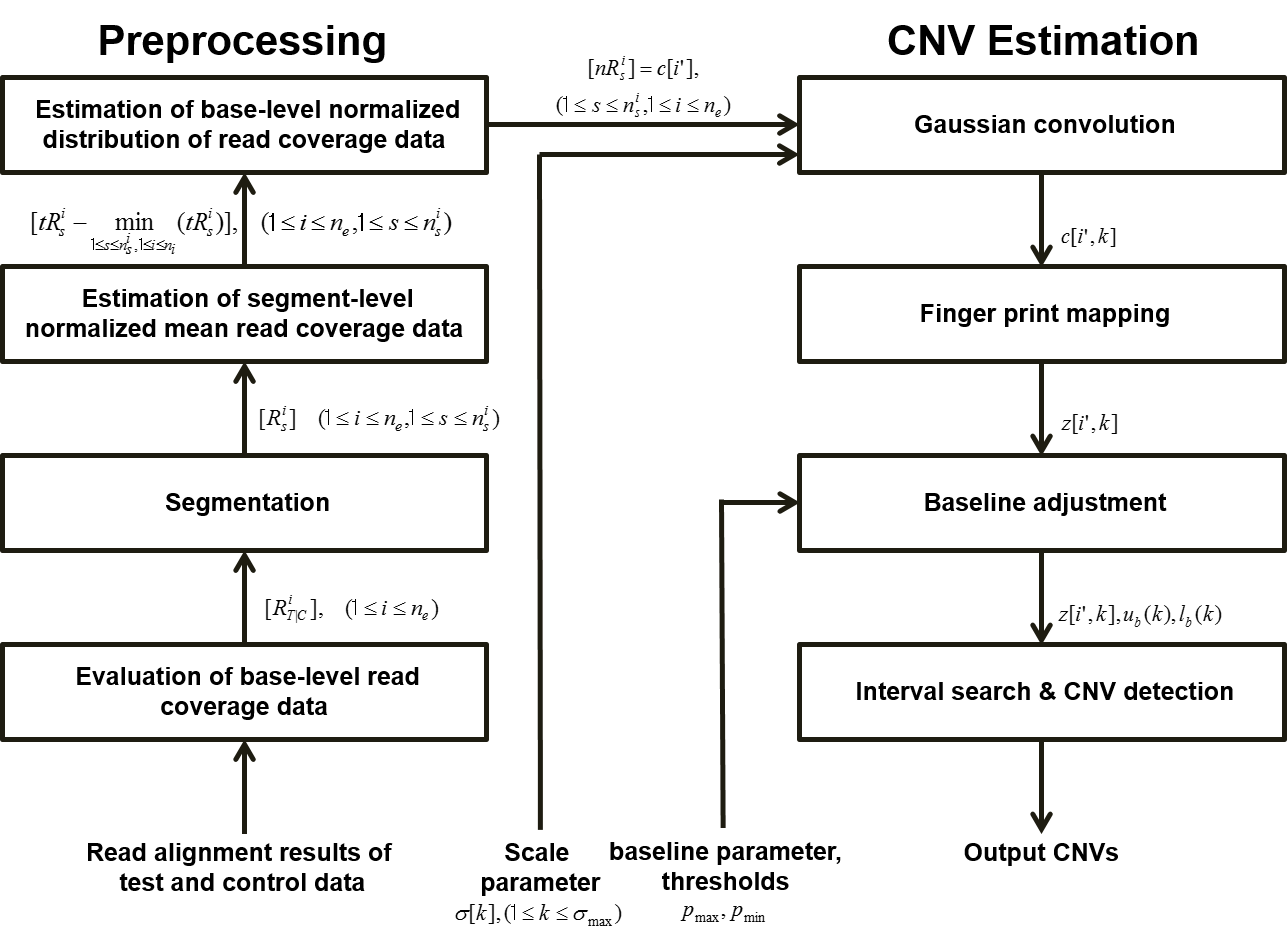
ExCNVSS flowchart: It includes two procedures: data preprocessing and CNV estimation. The data preprocessing procedure included a four-step normalization protocol. The CNV estimation procedure included a Gaussian convolution, finger print mapping, baseline adjustment, interval search and CNV detection.
|
|
