|
|
| How to run the PhenGenVar Browser |
|
1. Download the PhenGenVar Browser setup file and sample data from the Download page 2. Execute the setup file 3. Click the Next button .... 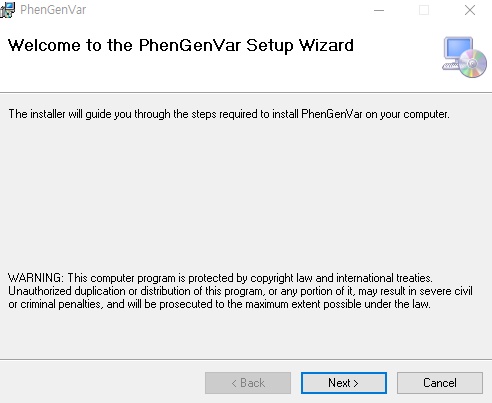 4. Select the installation folder and click the Next button .... 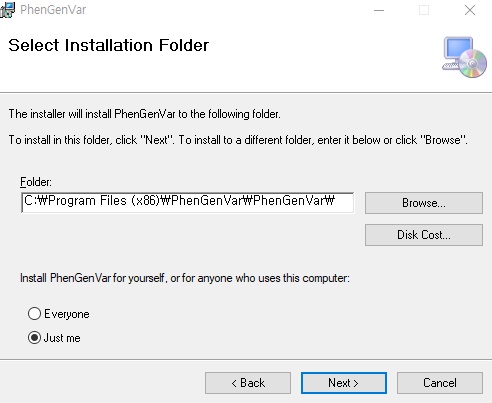 5. Click the Next button to start the installation .... 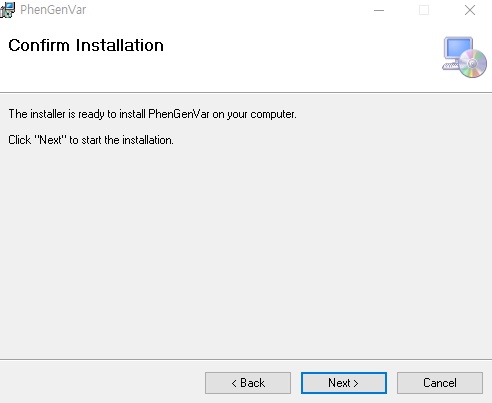 6. After the installation is successfully completed, click the Close button to exit .... 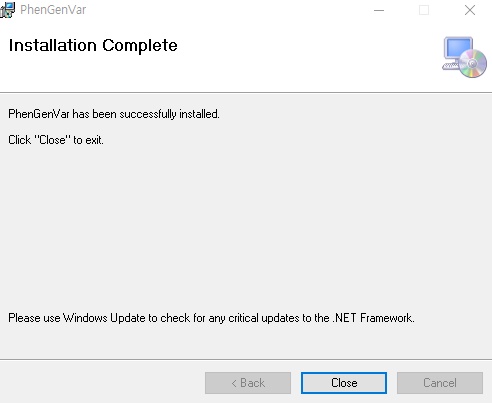 7. See the install icon on the desktop and double-click it to run the program 8. Unzip the sample data ....A. Sample read data : bam file ....B. Sample variant data : vcf file 9. Main Page of PhenGenVar Exome browser .... 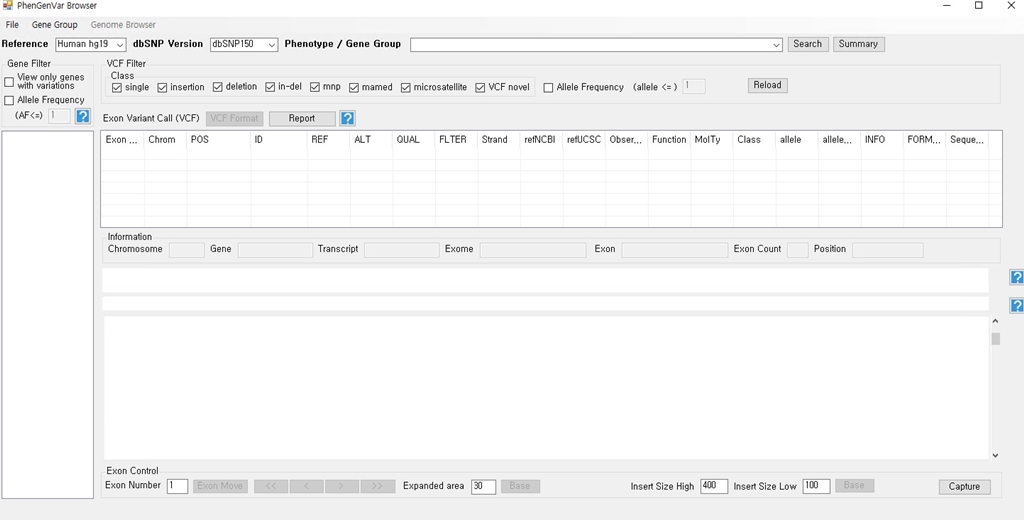 10. Select a Gene_Group or Phenotype to analyze and click the Search button .... 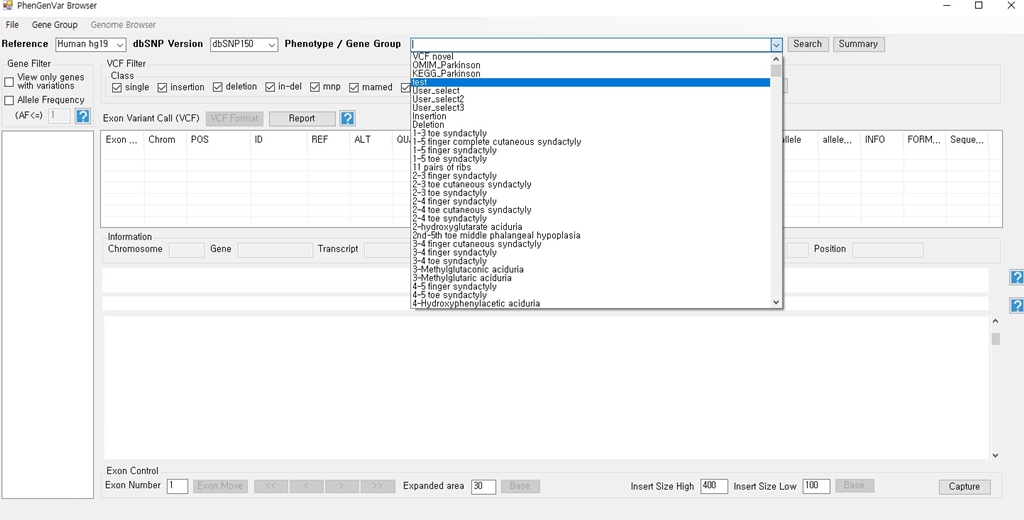 11. Select and doucle-click a random gene/transcript to analyze .... 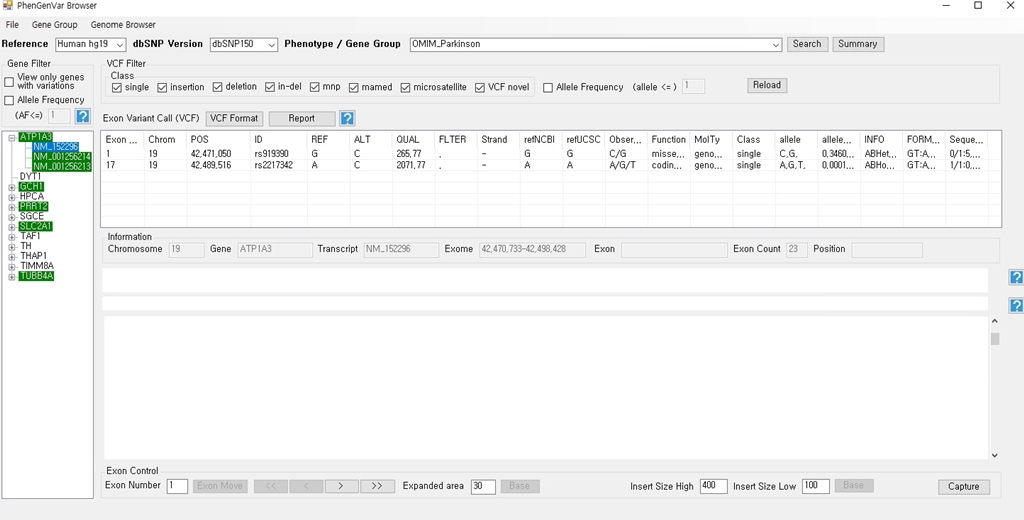 12. Select and click a random variant and analyze the genetic information of the variant .... 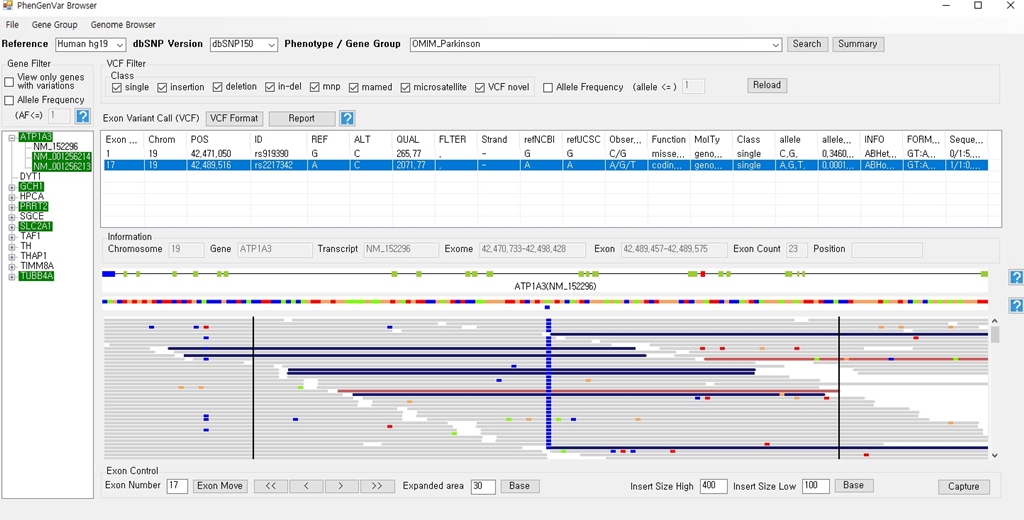 13. Double-click a random variant to call a Genome Browser and use it for a detailed analysis of that area .... 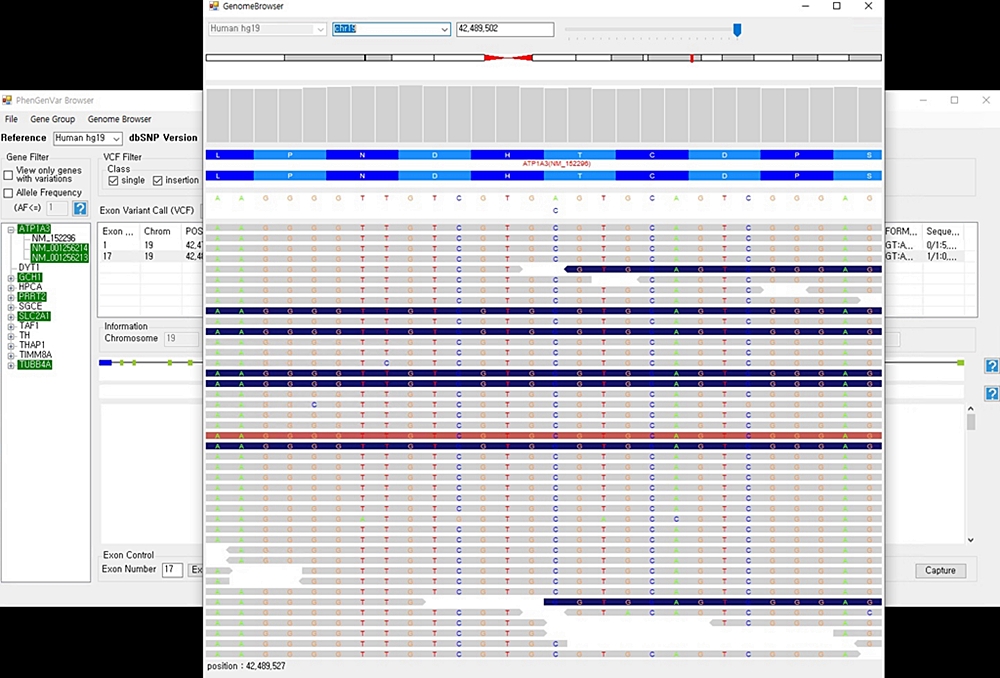 ※Refer to the manuals for detailed information on how to use the PhenGenVar Exome/Genome Browser.※ |
| Database Laboratory, Department of Computer Engineering, Hallym University, 1 Hallymdaehak-gil, Chuncheon, Gangwon-do 200-702, Korea |